
Rapprocher les modèles informatiques et les expériences in vivo pour comprendre l’évolution des bactéries
Two teams from Rhône-Alpes, including one from the LIRIS (Beagle Team - CNRS/INRIA/INSA-Lyon/UCBL), have just published a review in Nature Reviews Microbiology to show how recent developments on experimental evolution in vivo as well as in silico enable researchers to better understand bacterial systems and their evolutionary dynamics.
Experimental evolution is a recent experimental approach in evolutionary biology. In such experiments, organisms are let evolve in laboratory, in a perfectly controlled environment, during thousands or tens of thousands generations. Experimental evolution enables researchers to observe evolution "in action" to decipher the mechanisms at work during bacteria evolution. In this article, computer scientists from the LIRIS and biologists from the LAPM show how in vivo experiments can be confronted to in silico experiments (i.e. computational models of evolution) to identify generic evolutionary mechanisms. In a near future, co-development of in vivo and in silico experiments will enable to discover the key evolutionary parameters that impact the evolutionary fate of a bacterial population and the corresponding mechanisms at the ecological, cellular and molecular levels.
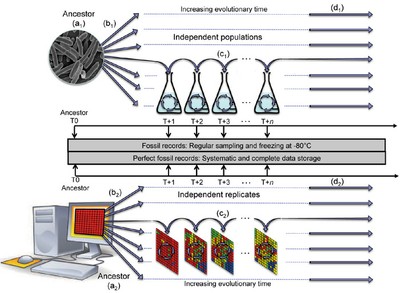
In vivo microbial and in silico evolution experiments. Ancestral microbial (top) or artificial organisms (bottom) are propagated in defined wet or computer environments, respectively. The main advantage is the ability to compare the "ancestor” with evolved populations sampled throughout evolution. All living and artificial organisms can be kept frozen or stored in database, respectively, and revived at any time for further analyses. Many parameters can be varied, including ancestors strain (a1, a2), number of independent replicates (b1, b2), population size and culture conditions (c1, c2) or duration of the experiments (d1, d2) ...
Reference :
New insights into bacterial adaptation through in vivo and in silico experimental evolution, Nature Reviews Microbiology 10, 352-365 (May 2012) | doi:10.1038/nrmicro2750
